What if we told you which cancer treatments could work best for you or a loved one?
Recent studies in genomics have made a step towards personalized care for cancer patients. Loss of essential genes may limit the effectiveness of treatments that activate the immune system to fight cancer.
On average 617 Canadians are diagnosed with cancer every day and undergo treatments including not only chemotherapy and radiation, but also immunotherapy. The immune system is our body’s mechanism to help fight pathogens that make us sick. It works when we fall off our bikes and scrape our knees; it also works when we fight illnesses, such as cancer. Immunotherapies are a type of treatment that boosts the immune system to help it battle cancer.
Cancer presents a challenge as it tends to avoid the immune system. Immunotherapies help strengthen our immune system so it can find cancer cells and destroy them. The downside to immunotherapies is that they don’t always work for everyone. But how do we know who they will work for? We can’t know for sure but looking at the genes in cancer cells can give us a hint.
Pathogens have DNA sequences that differ from our own, making it easy for the immune system to identify them, but cancer cells are an anomaly closely resembling human cells. They mutate their DNA to help them hide from the immune system and continue to grow excessively, resulting in tumours, which destroy surrounding tissues. To do so, cancer cells tend to “turn off” certain genes so that they can’t do their job right anymore—we call these loss-of-function mutations. Some of these mutations also decrease the effectiveness of immunotherapies. Knowing whether a tumour has mutations in these genes gives us a clue about how effective immunotherapy will be.
A recent study by Patel et al., discovered that the genes essential for effective immunotherapy usually have something to do with the function of killer T cells (CD8+ T cells). As their name suggests, these T cells are involved in killing cells infected by pathogens, and also cancer cells. Killer T cells operate by releasing lytic granules that tear down the target cell. Moreover, killer T cells release cytokines (signalling molecules) that help in mounting an immune response and clearing the pathogen. The study expanded even more so, identifying several specific gene mutations that are associated with survival rates of cancer patients treated with immunotherapies.
To do this, the authors used CRISPR-Cas9, a complex of molecules that acts like a pair of scissors and cuts DNA at specific sites, mimicking loss-of-function mutations in cancer cells (see below). If lytic granules and cytokines were not released, the function of the killer T cell was lost. This means the gene targeted by CRISPR-Cas9 is important in the T cell immune response. The genes in the cancer cells that are required for the production of cytokines by killer T cells were referred to as candidate genes.
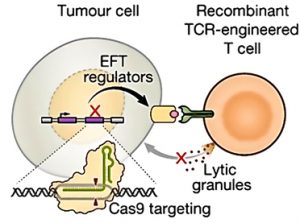
Adapted from Patel et al., 2017
CRISPR-Cas9 was used in tumour cells to identify which genes may be necessary to elicit the effector function of T cells (EFT). CRISPR-Cas9 essentially turned off certain genes to determine whether lytic granules, which cause cell death, would be released leading to lysis of tumour cells (see Patel et al., 2017).
After identifying which genes are important for killer T cell function, the authors needed to determine which of these genes are mutated in cancer patients. By doing so, patients’ receptiveness to immunotherapies can be determined by sequencing their tumour cells. They used DNA profiles of over 11 thousand tumour cells from The Cancer Genome Atlas (TCGA) that included 36 different cancer types. When compared with the candidate genes, the authors found 19 genes that were also mutated in cancer cells (see below). Some of these genes were B2M, TAP1/2 and JAK2. Loss-of-function mutations in these genes make it difficult for our immune system to recognize the cancer cells. Ultimately, this means that such mutations allow cancer cells to continue to grow into tumours, avoiding the immune system and reducing the effectiveness of immunotherapies.
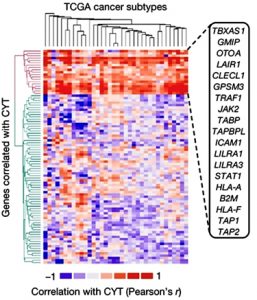
Adapted from Patel et al., 2017
Heatmap of gene clusters that are important for the cytolytic activity of killer T cells (CYT). In the y-axis, we see the candidate genes that are correlated with cytolytic activity. The x-axis shows the different subtypes of cancer mentioned in The Cancer Genome Atlas. If any of the highlighted genes are lost in tumour cells, there may be decreased or eliminated expression of tumour antigens to T cells, leading to immune evasion (see Patel et al., 2017)
Though the discovery of these 19 genes is outstanding, this post just scratches the surface of the results published by the authors. In addition to this, the authors delved into the response of a specific cytokine—interferon-γ—as well as the importance of microRNAs, which play a role in gene expression.
All in all, this study brings us one step closer to delivering personalized care for cancer patients. Knowing that a tumour may be receptive to immunotherapy means patients might not need to undergo chemotherapy with the side effects that accompany it.
Learn more about immunotherapy and genomics:
About the Author
This post was written by Iman Kassam. Currently, she is completing her undergraduate degree, majoring in molecular biology and mental health. Aside from academia, Iman is working at DNALabs Canada, where she researches how genetics affects health. After the completion of her degree, she hopes to spend time traveling before pursuing a career in health care! For now, Iman has been spending her time exploring trails close to home in the Canadian Rocky Mountains!
